MEETING HIGHLIGHT
The PROfound promise of genomics in mCRPC management
At the Uro-Oncology Asia 2021 Conference organized by the Hong Kong Society of Uro-Oncology, Dr. Joaquin Mateo, medical oncologist and attending physician at the Vall d’Hebron University Hospital, Barcelona, and principal investigator at the Vall d’Hebron Institute of Oncology, discussed the importance of genomics and precision medicine in the context of metastatic castration-resistant prostate cancer (mCRPC). As lethal prostate cancer has gene alterations that are distinct from indolent prostate cancer, and metastatic prostate cancer patients have enriched loss of genes encoding tumor suppressors and DNA repair, molecular stratification of the disease may help guide treatment selection based on genomic biomarkers. The PROfound trial is one study that found a significantly longer median radiographic progression-free survival (PFS) among olaparib treated patients when compared to enzalutamide/ abiraterone, which was even longer when only accounting for cohort A of the study that included patients with alterations at the BRCA1, BRCA2 or ATM gene.1 At the end, Dr. Mateo also discussed the practicalities of genomic testing in the clinical setting.
Progression of castration-resistant prostate cancer is associated with poor survival outcomes
Castration-resistant prostate cancer (CRPC) is an advanced cancer marked by disease progression following surgical or pharmaceutical (androgen deprivation) castration.2 Nearly all men with metastatic prostate cancer treated with primary androgen deprivation therapy will develop resistance, resulting in CRPC.3 According to the data from a systematic review of observational studies, approximately 10-20% of prostate cancer cases develop CRPC within 5 years of follow-up.2 In fact, more than 83% of patients have metastases at the time of CRPC diagnosis, and 33% without metastases at diagnosis will develop metastases within 2 years.2 As the median overall survival (OS) of mCRPC patients are known to be much shorter than nonmetastatic patients, methods to better stratify these metastatic patients based on their underlying risks would be important to guide treatment selection.
Genomic profiling can improve treatment selection and clinical outcome prediction
Genomic profiling, in addition to functional imaging for ascertaining tumor stage and blood tests for determining prostate-specific antigen evolution, is one such method that to complement the treatment decision-making process. In addition to identifying family risk, genomic markers can also be used to optimize therapy sequence and monitor drug resistance.4 However, the acquisition of satisfactory metastatic biopsy samples remains a challenge, and whether primary tumor samples can be used for stratification is still unclear.5
Nonetheless, insights on gene mutations unique to metastatic prostate cancer tumors can help clinicians select the optimal therapy for each patient. Approximately 30% of advanced prostate cancers contain genetic alterations, including TP53, AR, PTEN, RB1, FOXA1, APC and BRCA2, that might help predict responses to existing drugs.4,6 With cellular pathways such as PI3K, DNA repair, cell cycle, WNT/CTNNB1 and epigenetic regulators being significantly more frequently altered among lethal prostate cancers, drugs that target actionable DNA damage response alterations, including ataxia-telangiectasia and Rad3 related (ATR) inhibitors for ATM mutations and immune checkpoint blockade for MMR mutations, can also be used after understanding the underlying genomic profiles of the prostate cancer patient.6
In addition to differentiating lethal and indolent prostate cancers, genomic profiling can also be used to predict treatment outcomes based on the patients' genomic alterations. In one study that investigated 128 mCRPC patients treated with abiraterone or enzalutamide, 18 recurrent genomic alterations were identified, but only the tumor suppressor RB1 gene alteration was significantly associated with poor survival.7 On the other hand, RB1, AR and TP53 alterations were associated with shorter treatment duration that was likely due to treatment resistance leading to termination.7 Furthermore, RB1 and TP53 alterations were sometimes associated with neuroendocrine-like histology, which in turn conferred more aggressive clinical behavior.7 Due to the strong association between RB1 alteration and poor clinical outcome as well as resistance to androgen receptor therapies, early genomic profiling of prostate cancer patients can help identify those who will benefit the most from the currently available treatment options and provide timely care to those with specific gene alterations.
In particular, defects in DNA damage repair (DDR) genes, including BRCA2, CDK12 and ATM, were found to be more common in mCRPC when compared with nonmetastatic prostate cancer cohorts.5 For patients with metastatic hormone-naïve prostate cancer, early genomic profiling with primary prostate tumor biopsies can provide a rationale for synthetic lethal strategies such as poly (adenosine diphosphate-ribose) polymerase (PARP) inhibitors or platinum to incur maximum survival benefit in earlier stages of the disease.5 As such, genomic alterations, such as RB1 and DDR defects, should be profiled as early as the time of primary cancer diagnosis so as to better stratify prostate cancer patients according to their underlying genomic risks. “How do we decide which patients receive the more intense therapeutic approach upfront, versus those who may be able to deescalate the treatment after all, and avoid side effects? Also importantly, how do we monitor emerging drug resistance and anticipate disease progression so that we can deliver better care for our patients?”, reflected Dr. Mateo. Better qualification of genomic biomarkers will reveal more answers in the future.
Olaparib was associated with greater survival benefit among patients with BRCA alteration
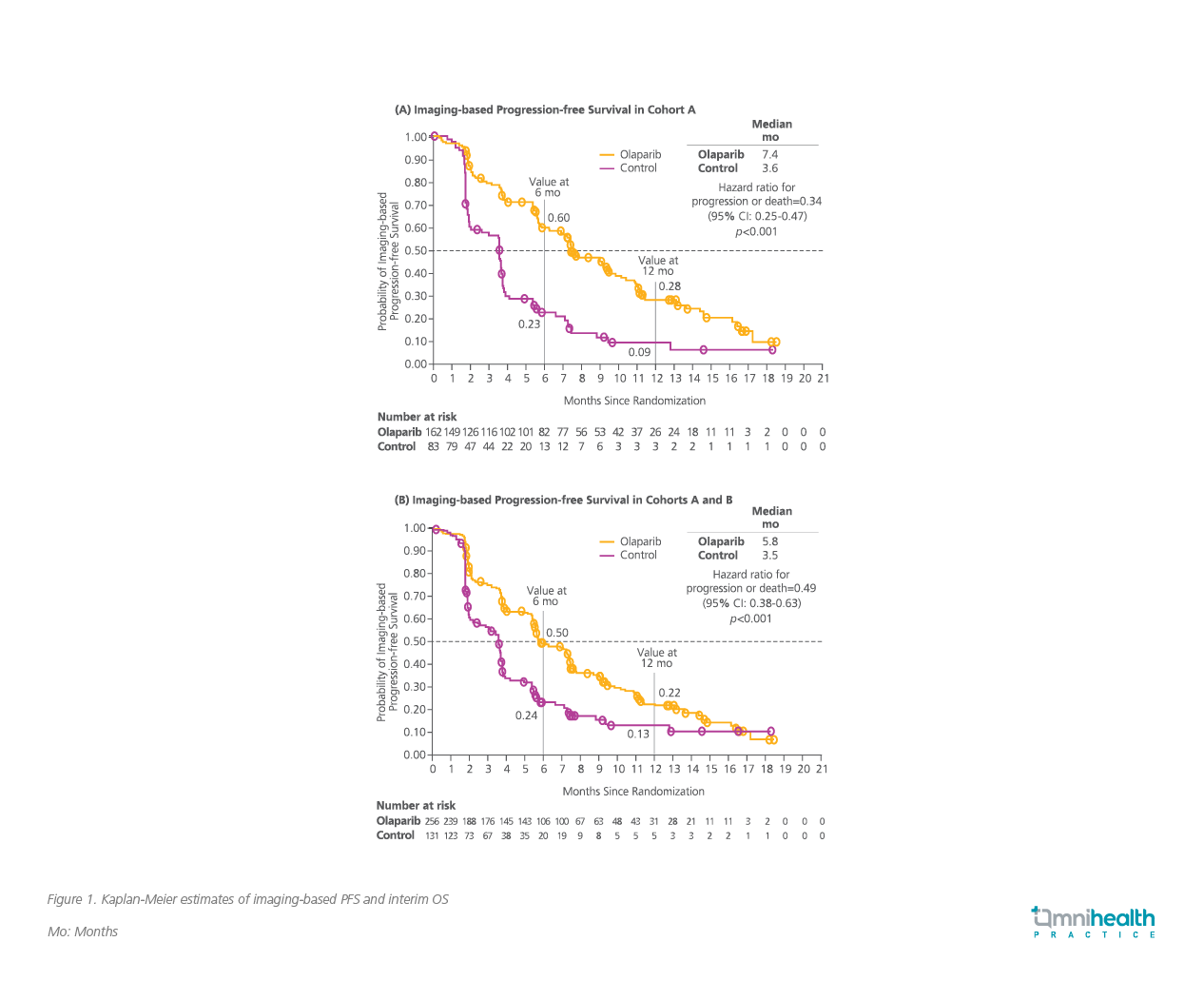
With an increasing number of gene alterations now identified as medically actionable, PARP inhibitors are the first example of a successful application of genomic biomarkers in the treatment of metastatic prostate cancer. To investigate the PARP inhibitor, olaparib, in men with mCRPC who had radiographic disease progression while receiving a new hormonal agent, the PROfound trial enrolled 387 patients who had alterations in ≥1 of any 15 qualifying genes with a direct or indirect role in homologous recombination repair.1 Patients were divided into 2 cohorts and were randomized to receive either a standard dose of olaparib or the physician’s choice of enzalutamide or abiraterone (control group); cohort A (n=245) had at least one alteration in the more commonly affected genes BRCA1, BRCA2 or ATM, while cohort B (n=142) had alterations in any of 12 other prespecified genes.1 The primary endpoint was imaging-based PFS in cohort A.1
Compared to the control group, olaparib significantly reduced radiographic cancer progression or death in cohort A by 66% (median PFS=7.4 vs. 3.6 months; HR=0.34, 95% CI: 0.25-0.47, p<0.001) (Figure 1A).1 In addition, olaparib significantly improved median OS in cohort A when compared to control (median OS=19.1 vs. 14.7 months; HR=0.69, 95% CI: 0.50-0.97, p=0.0175) after a median follow-up duration of 21.9 months in the olaparib arm and 21.0 months in the control arm.8 In the key secondary endpoint of PFS among the overall cohort (A+B), olaparib also demonstrated significantly longer median PFS when compared to control (5.8 vs. 3.5; HR=0.49, 95% CI: 0.38-0.63, p<0.001) (Figure 1B).1 Survival benefit was attained regardless of whether olaparib was administered before or after chemotherapy.
When considering the impact of particular gene alterations, an exploratory gene analysis revealed that patients with BRCA2 alterations received a greater PFS benefit with olaparib over the control (PFS=10.8 vs. 3.5 months) when compared to other DDR defects such as CDK12 (PFS=5.1 vs. 2.2 months) and ATM (PFS=5.4 vs. 4.7 months).1 Dr. Mateo elaborated, ”Major benefit [in survival] was seen among patients with BRCA1 and BRCA2 alterations. For ATM and CDK12 mutations, olaparib seemed to show some benefits but was quite minor compared to the benefits in the BRCA population.”
In terms of safety, the incidence of grade 3 or higher adverse events was higher with olaparib (51%) than with the control (38%).1 However, the duration of therapy in the olaparib group was nearly twice longer than the control group, which may be the reason contributing to a higher incidence of certain adverse events.1 Overall, the safety profile of olaparib was similar to that described in other monotherapy studies.1
Clinical translation of genomic testing
The PROfound study is just one illustration of how genomics can be used to guide patient decision-making. Recently, there has been a change in paradigm and Dr. Mateo pointed out that an update of various international guidelines now recommends that all patients with metastatic prostate cancer should be offered genomic sequencing for treatment optimization and identification of germline mutations.9–11 For instance, the National Comprehensive Cancer Network now recommends tumor testing of somatic homologous recombination gene mutations in patients with regional or metastatic prostate cancer.11
However, before these guidelines can be more widely adopted, potential technical problems such as inadequate tumor content, low DNA yield and DNA degradation due to prolonged storage as observed in the PROfound trial should be addressed. To overcome these challenges, Dr. Mateo advocated collecting sufficient material during biopsy, labeling the paraffin blocks individually, optimizing protocols for sampling handling and fixation, ensuring proper storage, performing quality control of samples early and streamlining the consent process and genetic counseling for patients. These additional steps will help improve the DNA yield and quality that eventually lead to higher accuracy in genomic profiling for better therapy selection.
Finally, Dr. Mateo cautioned that 20-40% of primary prostate cancer biopsies might fail next-generation sequencing tests due to technical complications. Metastatic biopsies are particularly challenging due to the decalcification in bone biopsies and anatomical difficulties in certain areas such as the retroperitoneal and pelvic nodes. Liquid biopsies are promising but also challenging as tumor DNA is diluted into non-tumoral circulating DNA in the blood, making the detection of deletions, such as those in BRCA2, particularly difficult.
Conclusion
Genomic testing is recommended and is increasingly being performed on men with metastatic prostate cancer. Certain genes are altered in lethal metastatic cancer compared to indolent cancer, with the RB1 gene being associated with poor survival. Genes such as BRCA1 and BRCA2 are particularly sensitive to PARP inhibition, and patients with such gene alterations treated with olaparib have demonstrated prolonged survival when compared with conventional androgen receptor inhibitors. In order to better select therapies and monitor potential treatment resistance, techniques to improve the DNA yield and quality should be adopted so that the goal of using prostate cancer genomics to improve therapy outcomes can be realized.

